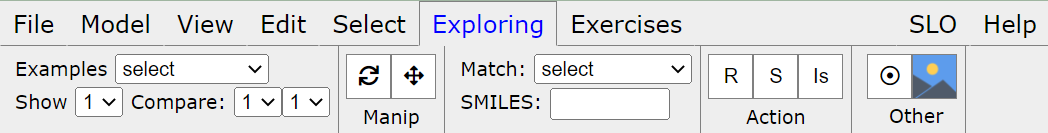
Click on the menu Exploring, submenu Comparisons for toolbar display

You can start with the currently loaded models (at least 2) or one of the examples given in the Examples list on the toolbar. You can reload the current example using R button.
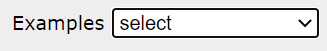
You can display all models at once (default) or each model separately using the Show list.

Models can overlap in the beginning. It is not necessary, but advisable to position the models side by side.

Clicking the arrows button and dragging a model with the mouse, you can spread the models manually.
You can rotate each model separately by clicking the rotate button and then dragging a model with the mouse.
You can zoom the view with the mouse wheel.

You can center the models using the center button.
Identifying common substructure. At least one common substructure has to be identified all currently displayed models. Substructure can be selected from a Match list or entered in the SMILES notation in the SMILES field.
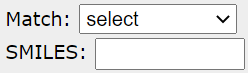
Identified substructure is then colored red in all the models.
Orienting the models. Using the R button, models are automatically rotated in place so that the common substructure is oriented the same way as in the first model.
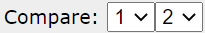
Some operations can only be done on two models at once.
Model pair selection. This is done in the compare list, where models are numbered. The first two models are selected by default. In case of more than 2 models other pairs can be selected.
Type of isomerism. Clicking on button Is automatically calculates the type of isomerism between the selected models. Results include: a) constitutional isomers, b) enantiomers, c) diastereomers, d) identical, e) none. No distinction is made among the different types of constitutional isomers.
Model superposition. A step further (or separate) from the model orientation. Button S animates superposition of the second model chosen on the list to the first model and colors it. This can be repeated for the next model.