


|
|||||||||||||

|
|||||||||||||
| Professor Dr Franc Gubensek | |||||||||||||
| Dr Dusan Kordis | |||||||||||||
by Dusan Kordis and Franc Gubensek
Ammodytin L is a myotoxic Ser49
phospholipase A2 (PLA2) homologue, which is tissue specifically
expressed in the venom glands of Vipera ammodytes. The complete
DNA sequence of the gene and its 5' and 3' flanking regions has
been determined. The gene consists of five exons separated by
four introns. Comparative analysis of the ammodytin L and ammodytoxin
C genes shows that all intron and flanking sequences are considerably
more conserved (93-97 %) than the mature protein-coding exons.
The pattern of nucleotide substitutions in protein-coding exons
is not random but occurs preferentially on the first and the second
positions of codons, which suggests positive Darwinian evolution
for a new function. A Ruminantia specific ART-2 retroposon, recently
recognised as a 5'-truncated Bov-B long interspersed repeated
DNA (LINE) sequence, was identified in the fourth intron of both
genes. This result suggests that ammodytin L and ammodytoxin C
genes are derived by duplication of a common ancestral gene. The
phylogenetic distribution of Bov-B LINE among vertebrate classes
shows that, in addition to the Ruminantia, it is limited to Viperidae
snakes (Vipera ammodytes, Vipera palaestinae, Echis coloratus,
Bothrops alternatus, Trimeresurus flavoviridis and Trimeresurus
gramineus). The copy number of the 3' end of Bov-B LINE in the
Vipera ammodytes genome is between 62 000 and 75 000. The absence
of Bov-B LINE at orthologous positions in other snake PLA2 genes
indicates that its retrotransposition in the V. ammodytes PLA2
gene locus has occurred quite recently, about 5 My ago. The amplification
of Bov-B LINEs in snakes may have occurred before the divergence
of the Viperinae and Crotalinae subfamilies. Due to its wide distribution
in Viperidae snakes, it may be a valuable phylogenetic marker.
The neighbor-joining phylogenetic tree shows two clusters of truncated
Bov-B LINE, a Bovidae and a snake cluster, indicating an early
horizontal transfer of this transposable element.
Keywords : ammodytin L, Bov-B long interspersed repeated DNA,
ART-2 retroposon, Viperidae, molecular evolution.
| Igor Krizaj, Ph.D, Natasa Vucemilo B.Sc and Alenka Eopie B.Sc |

|
During evolution many snake
venom phospholipases A2 (PLA2) [1] have acquired different physiological
activities, including presynaptic and postsynaptic neurotoxicity,
myotoxicity, blood-clotting activity, blood-pressure-depressing
activity [2], several of which can be present in a single species.
In the long-nosed viper (Vipera ammodytes), at least five different
PLA2 are present [3]. Ammodytin L (amd L) is the only natural
mutant of the group II PLA2, in which the active site Asp49, responsible
for the binding of Ca2+, is replaced by serine [4]. The ammodytoxin
C (amtx C) gene [5] has the same structure as other known mammalian
group II PLA2 genes [6] having 5 exons and 4 introns but differs
in structure from PLA2 genes of Crotalinae species [7, 8] all
of which show up to 90 % similarity in intron and flanking sequences.
In the ammodytoxin C gene, the highly conserved ART-2 retroposon
was found in its fourth intron [9]. This unusual occurrence was
explained by horizontal transfer of this transposable element
between vertebrate classes, a tick being a possible carrier. The
possible origin of ART-2 retroposon was erroneously ascribed to
U5 snRNA on the basis of incorrect GenBank data. The MUSUR5E sequence
[10] bears no relation to any other authentic U5 snRNA and could
have been reverse transcribed from contaminating bovine DNA [11].
At that time ART-2 (truncated Bov-B LINE) was still believed to
be a short interspersed repeated DNA (SINE), posing the problem
of how such a short, non-coding element could amplify in a newly
invaded genome [12]. The discovery of the Bov-B LINE in V. ammodytes
and other snake genomes is of considerable interest because this
provides the first evidence of horizontal relationships of LINEs
in vertebrates [9, 12].
ART-2 retroposons were independently discovered in Bovidae genomes
by Duncan [13] and Majewska et al. [14] and designated as ART-2
and Pst repetitive elements, respectively. Lenstra et al. [15]
renamed these repeats as Bov-B SINE elements. Here we use the
originally proposed term ART-2 retroposon when we refer to previous
work. Jobse et al. [16] studied the history of the Bov-B SINE
elements by comparative hybridisation and PCR, and found that
they emerged just after the divergence of the Camelidae and the
true ruminants. Recently, Modi et al. [17] used Southern blot
hybridisation and fluorescent in situ hybridisation (FISH) to
study the distribution of ART-2 retroposon in 46 species of artiodactyls,
and found that it is specific for all pecoran ruminants (fam.
Bovidae, Antilocapridae, Cervidae and Giraffidae). From both articles
it is clear that Bov-B SINEs have been found only in suborder
Ruminantia. FISH studies indicated that the ART-2 retroposons
are fairly evenly distributed among GTG-light and GTG-dark bands
and that this arrangement probably existed in the common ancestor
to pecoran ruminants.
Szemraj et al. [18] described a family of bovine 3.1-kb repetitive
sequences called the bovine dimer-driven family (BDDF), which
contains the complete ART-2 sequence at its 3' end. BDDF members
are mutated or truncated LINE-like elements encoding their own
reverse transcriptase. Szemraj proposed that the ART-2 retroposons
should be considered as truncated BDDF (LINE-like) elements. Copy
number estimates of the ART-2 retroposon in the bovine genome
range from 50 000 [14] to 200 000 [17] copies/genome.
Here we present the complete structure of ammodytin L gene and
observation of the distribution of truncated Bov-B LINE elements
in the genomes of other Viperidae snakes.
Screening of V. ammodytes genomic library. A V. ammodytes genomic library in the l GEM-12 [5] was screened with the ammodytin L cDNA [19] labeled with [35S] dCTP[S] by the random-priming method [20] using the plaque hybridisation method. Hybridisation was carried out at 42oC for 20 h in a mixture of 6 x NaCl/Cit (NaCl/Cit is 0.15 M NaCl and 0.015 M sodium citrate, pH 7.0), 5 x Denhardt's solution, 0.5 % SDS and denatured herring sperm DNA at 100 ?g/ml in 50% formamide. The filters were washed successively with 6 x NaCl/Cit and 2 x NaCl/Cit at 35 oC for 20 min each. The positive clones were rescreened by the same procedure.
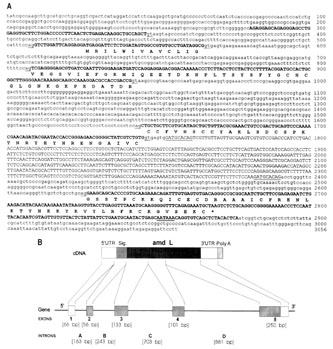
|
| Fig.1 Complete nucleotide sequence (A) and the structure (B) of the ammodytin L gene. The deduced amino acid sequence is presented below the coding parts of the exons. Splicing signals, signal for polyadenylation and direct repeats are underlined, exons are in bold capitals. Introns and both flanking regions are designated by small letters. Truncated Bov-B LINE, previously designated as ART-2 retroposon and positioned between direct repeats, is designated by capitals. The structure of the ammodytin L gene is compared with its cDNA. Five exons are indicated by boxes, the four introns and both flanking regions by lines. |
Characterisation of genomic clones. Phage DNA was prepared
from plate lysates [20] and digested with BamHI, EcoRI, SacI and
XhoI restriction enzymes. The resulting fragments were separated
by gel electrophoresis on 0.7 % agarose, transferred to Hybond-N
membranes (Amersham) and hybridised with the amd L cDNA probe
at 42oC as described above. Positive genomic fragments were subcloned
into pUC 19 (Pharmacia) and further digested with different restriction
enzymes. PstI and PstI-AvaI fragments were subcloned into pUC
19 by standard ligation and transformation techniques, using Escherichia
coli host strain DH 5a. Plasmid DNA was isolated by the method
of Sal et al. [21].
Copy number of truncated Bov-B LINE elements in V. ammodytes genome.
A V. ammodytes genomic library [5] was screened with the 628-bp
PstI fragment of ammodytin L gene containing the truncated Bov-B
LINE sequence, labeled with 32P by the random-priming method [20]
using the plaque-hybridisation method. Hybridisation conditions
were the same as described above.
DNA sequencing and analysis. Sequencing was carried out using
the dideoxy chain-termination method [22] with a T7 sequencing
kit following the supplier's protocol (Pharmacia). The nucleotide
sequence of both DNA strands was determined. Analysis of DNA sequences
was performed with the BLAST program [23] at NCBI. Nucleotide
sequences were cross-compared using the program CLUSTAL W [24].
Genomic DNA preparation and Southern blot analysis. Genomic DNA
was isolated from members of several vertebrate classes and an
invertebrate (tick) using standard procedures [20]. 10 ?g of PstI-digested
genomic DNA of each species was separated electrophoretically
in 1 % agarose gel and transferred to Hybond-N membranes (Amersham),
according to the supplier's recommendations. Membranes were hybridised
under the same conditions, as described above, with the probe
used for copy-number determination of truncated Bov-B LINE elements.
The final high-stringency wash was with 0.1 x NaCl/Cit plus 0.1
% SDS at 75oC.
Phylogenetic relationship of Bov-B LINE elements. Snake and some
Bovidae truncated Bov-B LINEs (cut to maximal overlapping length)
were aligned with the program CLUSTAL W [24]. Analyses were performed
on 500 alignment positions. Phylogenetic relationships were reconstructed
by the neighbor-joining method [25], with the Kimura two-parameter
model of distances, using program MEGA [26]. Tree reliability
was assessed by the bootstrap method, with 1000 replications using
program MEGA[26].
Isolation and sequencing of
ammodytin L genomic clone. The clone l 2.3, containing a 12.5-kb
insert, was isolated by screening the V. ammodytes genomic library
with a cDNA probe encoding the entire ammodytin L. It was characterised
by restriction analysis using BamHI, EcoRI, SacI and XhoI and
all possible combinations of pairs of these enzymes. After Southern
blot analysis, the positive genomic fragment carrying the complete
amd L gene was subcloned into the pUC 19 vector. It was further
digested with several restriction endonucleases. Only the PstI
and PstI-AvaI fragments, having the most suitable size, were subcloned
into pUC 19 for sequencing. The sequencing of larger segments,
containing introns, was completed using synthetic internal oligonucleotide
primers.
Structural organization of the ammodytin L gene. The complete
ammodytin L gene was found in a 3056 bp DNA segment (Fig 1A).
Alignment of the amd L cDNA and the genomic sequence demonstrates
that the amd L gene consists of a 5' flanking region followed
by 5 exons and 4 introns and a 3' flanking region (Fig 1b). Exon
1 encodes most of the 5'-UTR, exon 2 encodes the signal peptide
up to position -3, exons 3 to 5 encode the protein residues -3
to 42, 42 to 76 and 76 to 122 with the 3'-UTR, respectively. The
four introns of the amd L gene are located in positions homologous
to those occupied by the introns of the ammodytoxin C gene [5],
Crotalinae PLA2 genes [7, 8] and related mammalian group II PLA2
genes [6]. Within the coding region, introns B and D interrupt
the reading frame in phase I, and the intron C in phase II. The
5'-donor and 3'-acceptor splice sites in each of the introns conform
to the GT/AG rule [27]. The DNA sequences of all exons of the
ammodytin L gene are in agreement with the earlier published cDNA
sequence [19], except for 30 bp missing at the beginning of the
5' UTR of amd L cDNA in the latter sequence.
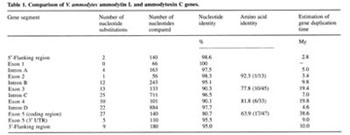
|
Comparison of the ammodytin L gene with the ammodytoxin C gene.
The sizes and nucleotide sequences of all four introns show a
high degree of conservation in both genes (Table 1) as well as
in other known PLA2 genes from the Crotalinae subfamily [5]. The
splice-site-encoded amino acids in the amd L gene are the same
as in the amtx C gene. The first two exons encoding the 5' UTR
and signal peptide are the most conserved exons in both genes.
Corresponding exons in each gene have the same size. The exons
coding for mature protein are much more divergent, especially
at the amino acid level. The high level of sequence identity of
the introns, the conservation of both flanking and untranslated
regions, and identical positions of truncated Bov-B LINEs in both
genes indicate that the latter are likely to have arisen by duplication
and divergence of a common ancestral gene. The pattern of nucleotide
substitutions in the coding regions, with most of the nucleotide
changes accounting for amino acid substitutions (Table 2), is
very unusual and indicates that these genes are under strong positive
Darwinian selection. The same was observed in Crotalinae PLA2
genes [7, 8]. Changes in protein-coding regions provided the snake
with a new pharmacological activity, which could increase the
effectiveness of the venom. The efficient mechanism of diversification
found in the PLA2 multigene family may have been needed to allow
rapid adaptation of the snakes for defense and for predation of
a wide spectrum of different prey - insects, fishes, amphibians,
reptiles and mammals.
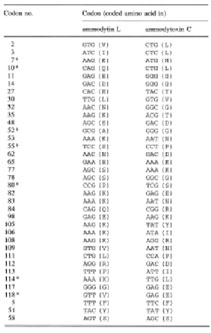
|
| Table 2. Nucleotide and amino acid (in parentheses) sequence differences between ammodytin L and ammodytoxin C mature protein coding regions. The last three codons (5, 51, 58) represent the only silent mutations. Asterisks denote the radical amino acid replacements, all other replacements are conservative |
Bov-B LINE elements in V. ammodytes PLA2 genes. Comparison of
highly conserved intron sequences of Viperidae venom PLA2 genes
[5, 7, 8] revealed that ammodytin L and ammodytoxin C genes contain,
in the fourth intron, a 630-bp long sequence which has not been
found in PLA2 genes of other Viperidae species. The sequence is
75 % identical to the consensus ART-2 retroposon sequence (accession
no. X82879) and shows approximately the same degree of similarity
to numerous ART-2 retroposons or truncated Bov-B LINE elements
from cattle (Bos taurus), goat (Capra hircus), sheep (Ovis aries)
and water buffalo (Bubalus arnee). Such a high level of similarity
undoubtedly shows their common evolutionary origin. The lengths
of the transposable elements in both PLA2 genes are almost the
same, they occur in the same position and differ in sequence by
only 2.4 %. An alignment of the truncated Bov-B LINE sequence
from the ammodytin L gene with that from ammodytoxin C, with the
consensus ART-2 sequence and with two shorter Bov-B LINE fragments
(283 and 288 bp long) found in the third intron of the TATA-box
binding protein (TBBP) genes in Trimeresurus flavoviridis and
Trimeresurus gramineus [28], is shown in Fig. 2. The sequences
from the three Viperidae species are nearly 90 % identical. The
finding of Bov-B LINE elements in T. flavoviridis and T. gramineus
genomes indicates that, in addition to ruminants, they may be
spread in Viperidae genomes.

|
| Fig. 2 Comparison of the truncated Bov-B LINE from amd L gene with that of amtx C gene, ART-2 consensus sequence (accession no. X82879) and related sequences from TATA -box binding protein (TBBP) genes from Trimeresurus flavoviridis (Tfl) and T. gramineus (Tgr). Alignment was constructed with the program Clustal W. The asterisks represent the nucleotides conserved between all sequences. |
The copy number of truncated Bov-B LINE elements in V. ammodytes
genome. Southern blot analysis has shown that truncated Bov-B
LINE elements are highly repeated in the V. ammodytes genome (Fig.
3). Their copy number was estimated by screening a l GEM-12 genomic
library with 32P-labeled truncated Bov-B LINE from amd L gene.
25 - 30 % of the plaques were positive. Assuming that the V. ammodytes
genome contains 3 x 109 bp/haploid genome and the average insert
size in l clones is about 12 kb, the copy number of 3' end of
Bov-B LINE elements can be estimated to be between 62 000 and
75 000 copies.
Phylogenetic distribution of Bov-B LINEs. The detection of truncated
Bov-B LINEs in the V. ammodytes genome, in addition to genomes
of ruminants, sheds new light on the present understanding of
the transmission and distribution of LINE elements [9, 12, 29].
Their presence in two vertebrate classes raises the question of
their distribution in other vertebrate classes and of their mode
of transmission between distant phylogenetic taxa. To examine
its possible presence in other vertebrate classes, a Southern
blot analysis was performed, using PstI-digested genomic DNA from
members of classes Mammalia (Homo sapiens, Sus scrofa, Canis familiaris,
Mus musculus and, as a positive control, Ovis aries and Capra
hircus), Aves (chicken Gallus sp.), Reptilia (Vipera ammodytes,
Vipera palaestinae, Echis coloratus, all Viperinae subfamily,
Bothrops alternatus from the Crotalinae subfamily, and Podarcis
muralis from the order Sauria) and Amphibia (Xenopus sp.). An
Arthropoda species (tick Ixodes ricinus), as a possible vector
[9], was also included. As a probe, a 32P-labeled truncated Bov-B
LINE from amd L gene was used.
In addition to V. ammodytes, this LINE was found in the genomes
of Viperinae (V. palaestinae, E. coloratus) and Crotalinae snakes
(B. alternatus, T. flavoviridis and T. gramineus). This may indicate
that its amplification in snakes occurred before the divergence
of Viperinae and Crotalinae subfamilies. The truncated Bov-B LINE
(ART-2 retroposon), originally ascribed to ruminants [13, 17],
has apparently a much wider phylogenetic distribution than previously
thought. The infiltration of Bov-B LINEs into the genomes of the
species examined, in two vertebrate classes, Reptilia and Mammalia,
may have occurred independently at approximately the same time
and presumably also by a common vector. Southern blot analysis
has also shown that in the Mammalia which tested, similar sequences
are not present outside the Bovidae family, neither are they present
in chicken, lizard, frog and tick genomes. Smit [12], in his recent
review, suggests that the invasion of Bov-B LINE elements has
occurred about 30 million years ago in the ruminant genome.
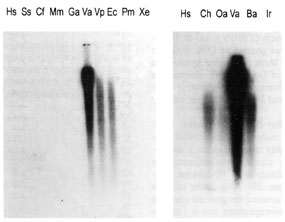
|
| Fig. 3 Phylogenetic distribution of truncated Bov-B LINE elements. Southern blot of PstI-digested genomic DNA from the members of different vertebrate classes and an invertebrate (tick) was hybridised with 32P labeled ART-2 probe. Genomic DNA samples from the following species were analyzed : human, Hs (Homo sapiens); pig, Ss (Sus scrofa); dog, Cf (Canis familiaris); mouse, Mm (Mus musculus); chicken, Ga (Gallus sp.); long-nosed viper, Va (Vipera ammodytes); Palestinian viper, Vp (Vipera palaestinae); Ec (Echis coloratus); Pm (Podarcis muralis); Xe (Xenopus sp.); goat, Ch (Capra hircus); sheep, Oa (Ovis aries); Ba (Bothrops alternatus); and a tick, Ir (Ixodes ricinus). |
Truncated Bov-B LINE elements in the V. ammodytes PLA2 gene locus
are very young. In order to estimate the time of integration of
the truncated Bov-B LINEs into V. ammodytes PLA2 gene locus, we
examined their distribution in several species of the Viperidae
family by Southern blot hybridisation and sequencing of V. ammodytes
PLA2 genes [30]. The low degree of divergence between truncated
Bov-B LINE elements and their limited presence in the fourth intron
of amtx C and amd L genes in V. ammodytes, but not in the orthologous
loci of other snake species, where they are abundant in the genomes,
indicates that retrotransposition into both PLA2 genes has occurred
very recently but before the gene duplication leading to amd L
and amtx C genes.
It is well known that recombination and/or gene conversion can
create situations where distinct regions of the same gene (exons,
introns, transposable elements) may have different evolutionary
histories. Because evolutionary rates differ significantly between
introns and exons (Table 1) in both PLA2 genes, this raises the
question as to which part of the gene can be used to infer divergence
times in the case of genes evolving under positive Darwinian selection.
From calculations of the divergence times of particular regions
in both genes (Table 1), it is evident that in the genes evolving
under positive Darwinian selection, the conserved introns may
be useful for the estimation of the age of duplicated genes. The
time of the insertion of the LINE in both PLA2 genes is estimated
at 4.8 My ago, using a 0.5 % nucleotide substitution/My [31].
This estimate is within the average range of time 6.5 My, inferred
from the intron and both flanking regions.
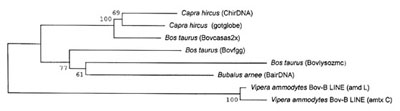
Phylogenetic relationships of Bov-B LINE elements. To clarify
the phylogenetic relationships between snake and Bovidae Bov-B
LINEs, we first aligned some sequences (Fig. 4) and then constructed
a phylogenetic tree using the neighbor-joining method [25] shown
in Fig. 5 where two distinct clusters, the Bovidae and Serpentes,
clearly separated. The grouping of these LINE elements from different
species or different genes from the same species is supported
with the high bootstrapping values as shown in Fig. 5.
Sequence analysis of the B. taurus Bov-B LINE elements (data not
shown) indicates that they belong to subfamilies of different
evolutionary ages, as has been observed for many other LINE elements
[32]. The V. ammodytes and B. taurus sequences display a degree
of similarity only slightly less than that displayed by the highly
divergent Bov-B LINE subfamilies in the bovine genome (75-95 %).
The transposable elements found in species that belong to different
genera, families, orders, classes and even kingdoms, may sometimes
be very similar. Such similarities are generally restricted to
a small part or parts of the nucleotide or protein sequences,
but are much greater than might be expected for species so distant
phylogenetically [33]. By contrast, the similarity of the truncated
Bov-B LINE elements among V. ammodytes and Bovidae is not restricted
to a small part of the nucleotide sequence, but is distributed
throughout the whole sequence without any gaps. Its presence in
two vertebrate classes, and the high level of similarity, suggests
that horizontal transfer is the only possible explanation of the
origin of this LINE. It is inconceivable that these sequences
could persist in non-coding regions of mammalian and reptilian
genomes which diverged over 250 My ago and still retain the present
level of similarity, particularly since, in most of the mammalian
LINE elements of different species, the similarity is limited
only to the coding regions, whereas in their 3' UTR, it has disappeared
[29, 34].
Possible carrier of Bov-B LINE elements. Although we proposed
Ixodes as a possible vector for the horizontal transmission of
the ART-2 retroposon between vertebrate classes [9] we did not
obtain any hybridisation signal in the Southern blot analysis
of the Ixodes genomic DNA tested. There are a few possible explanations
for this result. The sequences might be too divergent at the nucleotide
level to be identified by DNA hybridisation techniques, it might
not be present in the Ixodes genome, or Ixodes might not have
been involved in its transmission. As Capy et al. [33] pointed
out, however, it is not necessary for a vector to have the transposable
element integrated in its genome, it can instead operate as a
mechanical vector, as in the case of semiparasitic mite transfer
of the P-element between two Drosophila species [35, 36].
|
| Fig. 5 Phylogenetic relationships between Bov-B LINE elements. The neighbor-joining phylogenetic tree was based on the Clustal W multiple alignment from Fig. 4. To assess the reliability of branching patterns, 1000 bootstrap replications were performed. Numbers at the nodes indicate the bootstrap confidence level as a percentage. Sequence name abbreviations are from Fig. 4. |
LINE and SINE elements use similar mechanisms of retrotransposition,
which is similar to that of the R2 retrotransposon [37]. This
mechanism requires only the sequences at the extreme 3' end of
the RNA transcript, so that 5' truncated elements, which are truncated
Bov-B LINE elements, can still be integrated, and if these truncated
elements are transcribed they can generate new families of repeated
elements in a species. It seems clear that the boundary between
LINE and SINE elements will become more difficult to define in
the future. Bov-B LINE elements and LINE-derived parts of tRNA-SINE
elements [38] are presently the most striking examples of this
difficulty.
Bov-B LINE as a phylogenetic marker. The appearance of the Bov-B
LINE in V. ammodytes and other snake genomes is of considerable
interest because it is the first example of mammalian LINE-specific
elements observed in a species outside the class Mammalia [9].
The defective copies of LINEs, once inserted, appear to remain
stable in the genome of the host [29]. Comparisons between mammalian
b-globin loci have shown that different species can be distinguished
by the pattern of LINE-1 insertions at this site [39]. LINE and
LINE-like sequences have been found in every mammalian genome
studied and in a number of non-mammalian genomes as well, and
are species specific [29, 34]. The insertion of LINE sequences
without doubt affects the genomes, where they cause deletions
or duplications of regions by unequal crossover. Unfortunately,
snakes are among the least studied organisms in genetic terms.
It is thus premature to comment on the mutational impact of Bov-B
LINE elements in the snake genomes. It is possible that their
high copy number in closely related Viperinae and Crotalinae snake
species may have played a role in their speciation, perhaps by
facilitating reproductive isolation, as proposed for LINE-1 elements
in rodents [29].
Acknowledgement. For critical reading of the manuscript we thank
Prof. R. Pain. We are also indebted to Dr A. Smit for his valuable
comments. This work was supported by the Ministry of Science and
Technology of Slovenia by grant no.: P3-5243-0106.
| Gregor Anderluh, B.Sc and Loulou Kroon-Zitko, B.Sc |
|
References
1. Dennis,
E. A. (1994) Diversity of group types, regulation, and function
of phospholipase A2. J. Biol. Chem. 269, 13057-13060.
2. Kini, M. R. & Evans, H. J. (1989) A model to explain the
pharmacological effects of snake venom phospholipases A2. Toxicon
27, 613-635.
3. Gubensek, F, Ritonja, A., Zupan, J. & Turk, V. (1980) Basic
proteins of Vipera ammodytes venom. Basic studies. Period. Biol.
82, 443-447.
4. Krizaj, I., Bieber, A. L., Ritonja, A. & Guben1ek, F. (1991)
The primary structure of ammodytin L, a myotoxic phospholipase
A2 homologue from Vipera ammodytes venom. Eur. J. Biochem. 202,
1165-1168.
5. Kordis, D. & Gubensek, F. (1996) Ammodytoxin C gene helps
to elucidate the irregular structure of Crotalinae group II phospholipase
A2 genes. Eur. J. Biochem. 240, 83-89.
6. Seilhamer, J. J., Pruzanski, W., Vadas, P., Plant, S., Miller,
J. A., Kloss, J. & Johnson, L. K. (1989) Cloning and recombinant
expression of phospholipase A2 present in rheumatoid arthritic
synovial fluid. J. Biol. Chem. 264, 5335-5338.
7. John, T. R., Smith, L. A. & Kaiser, I. I. (1994) Genomic
sequences encoding the acidic and basic subunits of Mojave toxin
: unusually high sequence identity of non-coding regions. Gene
139, 229-234.
8. Nakashima, K., Nobuhisa, I., Deshimaru, M., Nakai, M., Ogawa,
T., Shimohigashi, Y., Fukumaki, Y., Hattori, M., Sakaki, Y., Hattori,
S. & Ohno, M. (1995) Accelerated evolution in the protein-coding
regions is universal in crotalinae snake venom gland phospholipase
A2 isozyme genes. Proc. Natl. Acad. Sci. USA 92, 5605-5609.
9. Kordis, D. & Gubensek, F. (1995) Horizontal SINE transfer
between vertebrate classes. Nature Genetics 10, 131-132.
10. Hamada, K., Kumazaki, T., Mizuno, K., & Yokoro, K. (1989)
A small nuclear RNA, U5, can transform cells in vitro. Mol. Cell.
Biol. 9, 4345-56.
11. Lenstra J. A. (1992) Bovine sequences in rodent DNA. Nucleic
Acids Res. 20, 2892.
12. Smit A. F. A (1996) The origin of interspersed repeats in
the human genome. Curr. Opin. Genet. Dev. 6, 743-748.
13. Duncan, C. H. (1987) Novel Alu-type repeats in artiodactyls.
Nucleic Acids Res. 15, 1340.
14. Majewska, K., Szemraj, J., Plucienniczak, G., Jaworski, J.
& Plucienniczak, A. (1988) A new family of dispersed, highly
repetitive sequences in bovine genome. Biochim. Biophys. Acta
949, 119-124.
15. Lenstra, J. A., Van Boxtel, J. A. F., Zwaagstra, K. A. &
Schwerin, M. (1993) Short interspersed nuclear element (SINE)
sequences of the Bovidae. Animal Genet. 24, 33-39.
16. Jobse, C., Buntjer, J. B., Haagsma, N., Breukelman, H. J.,
Beintema, J. J. & Lenstra, J. A. (1995) Evolution and recombination
of bovine DNA repeats. J. Mol. Evol. 41, 277-283.
17. Modi, W. S., Gallagher, D. S. & Womack, J. E. (1996) Evolutionary
histories of highly repeated DNA families among the Artiodactyla
(Mammalia). J. Mol. Evol. 42, 337-349.
18. Szemraj, J., Plucienniczak, G., Jaworski, J. & Plucienniczak,
A. (1995) Bovine Alu-like sequences mediate transposition of a
new site-specific retroelement. Gene 152, 261-264.
19. Pungerear, J., Liang, N. S., ©trukelj, B. & Guben1ek,
F. (1990) Nucleotide sequence of a cDNA encoding ammodytin L.
Nucleic Acids Res. 18, 4601.
20. Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular
Cloning: a laboratory manual, 2nd edn, Cold Spring Harbor Laboratory,
Cold Spring Harbor NY.
21. Sal, D. G., Manfioletti, G. & Schneider, C. (1988) A one
tube plasmid DNA mini preparation suitable for sequencing. Nucleic
Acids Res. 16, 9878.
22. Sanger, F., Nicklen, S. & Coulson, A. R. (1977) DNA sequencing
with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA
74, 5463-5467.
23. Altschul, S. F., Gish, W., Miller, E. W. & Lipman, D.
J. (1990) Basic local alignment search tool. J. Mol. Biol. 215,
403-410.
24. Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994)
CLUSTAL W : improving the sensitivity of progressive multiple
sequence alignment through sequence weighting, position-specific
gap penalties and weight matrix choice. Nucleic Acids Res. 22,
4673-4680.
25. Saitou, N. & Nei, M. (1987) The neighbor-joining method
: a new method for reconstructing phylogenetic trees. Mol. Biol.
Evol. 4, 406-425.
26. Kumar, S., Tamura, K. & Nei, M. (1994) MEGA : Molecular
evolutionary genetics analysis software for microcomputers. CABIOS
10, 189-191.
27. Senapathy, P., Shapiro, M. B. & Harris, N. L. (1990) Splice
junctions, branch point sites and exons : sequence statistics,
identification and application to genome project. Methods Enzymol.
183, 252-278.
28. Nakashima, K., Nobuhisa, I., Deshimaru, M., Nakai, M., Ogawa,
T., Shimohigashi, Y., Fukumaki, Y., Hattori, M., Sakaki, Y., Hattori,
S. & Ohno, M. (1995) Structures of genes encoding TATA box-binding
proteins from Trimeresurus gramineus and T. flavoviridis snakes.
Gene 152, 209-213.
29. Furano, A. V., & Usdin, K. (1996) DNA "fossils"
and phylogenetic analysis:using L1 (LINE-1, long interspersed
repeated) DNA to determine the evolutionary history of mammals.
J. Biol. Chem 270, 25301-25304.
30. Kordis, D. (1993) The structures of Vipera ammodytes PLA2
genes. PhD Thesis, University of Ljubljana.
31. Li, W-H., Gojobori, T. & Nei, M. (1981) Pseudogenes as
a paradigm of neutral evolution. Nature 292, 237-239.
32. Deininger, P. L., Batzer, M. A., Hutchinson ; III, C. A.,
& Edgell, M. H. (1992) Master genes in mammalian repetitive
DNA amplification. Trends Genet.8, 307-311.
33. Capy, P., Anxolabehere, D. & Langin, T. (1994) The strange
phylogenies of transposable elements : are horizontal transfers
the only explanation ?. Trends Genet. 10, 7-12.
34. Singer, M. & Skowronski, J. (1985) Making sense out of
LINEs : long interspersed repeat sequences in mammalian genomes.
Trends Biochem. Sci. 10, 119-122.
35. Houck, M. A., Clark, J. B., Peterson, K. R. & Kidwell,
M. G. (1991) Possible horizontal transfer of Drosophila genes
by the mite Proctolaelaps regalis. Science 253, 1125-1129.
36. Kidwell, M. G. (1993) Lateral transfer in natural populations
of eukaryotes. Annu. Rev. Genet. 27, 235-56.
37. Luan, D. D., Korman, M. H., Jakubczak, J. L., & Eickbush,
T. H. (1993) Reverse transcription of R2Bm RNA is primed by a
nick at the chromosomal target site : a mechanism for non-LTR
retrotransposons. Cell 72, 595-605.
38. Ohshima, K., Hamada, M., Terai, Y. & Okada, N. (1996)
The 3' ends of tRNA-derived short interspersed repetitive elements
are derived from the 3' ends of long interspersed repetitive elements.
Mol. Cell. Biol. 16, 3756-3764.
39. Hardison, R., & Miller, W. (1993) Use of long sequence
alignments to study the evolution and regulation of mammalian
globin gene cluster. Mol. Biol. Evol. 10, 73-102.